Automate, minimize the cost, and eliminate the use of pipette tips in scRNA (single cell RNA) NGS library prep protocols with I.DOT
With its unprecedented capacity for the analysis of large-scale genomic sequences, Next Generation Sequencing (NGS) is being adopted into a broad range of biological and biomedical applications. Rapid advances in NGS technologies and reduction in the cost of DNA sequencing have pushed its use into new territories: innovative research fields, increasingly larger study populations, and more and more complex
data analyses.
NGS is steadily integrated into clinical laboratory analysis, testing, and disease diagnostics in healthcare sectors worldwide. Further, NGS is gaining momentum as a means of choice for drug target identification efforts. "There are never two identical leaves," as the proverb goes, and this is true for the fundamental unit of life, the cell. Emerging data suggest that cells, even those generated from the same cell line or individual, have unique genomes, transcriptomes, and epigenomes due to genome and epigenome reprogramming, as well as DNA replication errors during cell division and differentiation.
Therefore, studies at the single-cell level are critical for understanding life processes, and their accomplishment has become the ultimate objective for those building biological technology. Low-throughput single-cell analysis techniques, such as immunofluorescence, single-cell PCR, and single-cell real-time PCR, have long been used to identify specific molecular markers of individual cells, but high-throughput profiling technology has been limited until recently.
With the improvement of next-generation sequencing technologies, single-cell sequencing methods have expanded, allowing for genome-wide screening of single cells at single-base resolution. There are several techniques for scRNA sequencing available now, including CEL-seq2, Drop-seq, MARS-seq, SCRB-seq, Smart-seq, and Smart-seq2 approaches. Many factors impact these procedures, and each method has its own unique workflow to generate sensitive, accurate, exact, and efficient
sequencing results; also, the strengths and limitations of many methods are previously recognized. However, the monetary cost of these solutions is still considerable and has not been adequately evaluated.
A recent study by Ziegenhain et al. (2017, Molecular Cell 65, 631–643) evaluated various aspects of six prominent scRNA-seq methods: CEL-seq2, Drop-seq, MARS-seq, SCRB-seq, Smart-seq, and Smart-seq2 for 583 mouse embryonic stem cells given a comprehensive insight into the benefits, drawbacks, and costs of the methods. The cost associated with scRNA library preparation range up to $25 (Table 1)
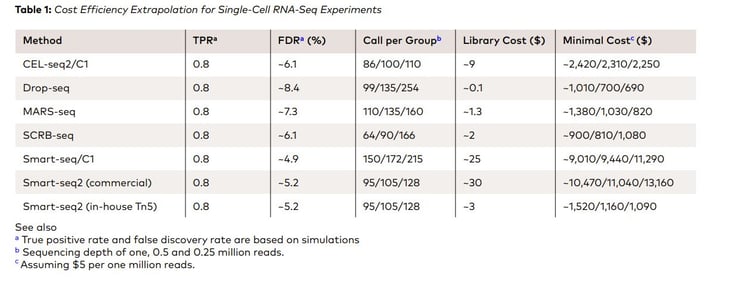
Similarly, AlJanahi et al. (2018, Methods & Clinical Development Vol. 10) evaluated widely used single-cell sequencing methods. According to the study, the most commonly used methods of library preparation cost up to $1.70 per cell (Table 2)
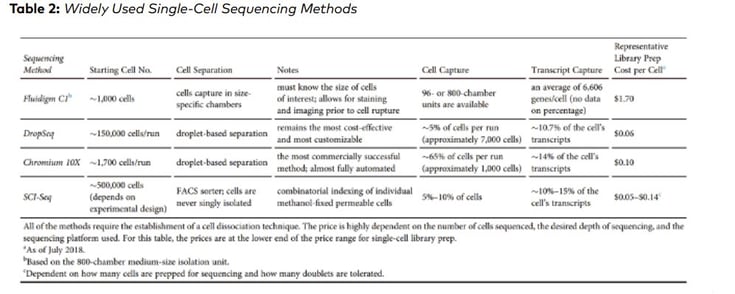
However, the given cost for library preparation does not include the consumable cost – for example pipette tips, required for library preparation protocol. Considering the costs associated with pipette tips would significantly increase the total cost for scRNA preparation.
The I.DOT in conjunction with plexWell™ Rapid Single Cell kit reduces costs, enhances throughput, and promotes sustainability. plexWell Rapid Single Cell for scRNA seq library Preparation from seqWell offers end-to-end library preparation for scalable, cost-effective single-cell transcriptome analysis in a single day, as well as a high-efficiency workflow that provides full-length transcript coverage with extremely sensitive transcript and gene detection.
Implementing I.DOT in scRNA library preparation workflow can reduce the cost expenditures on pipette tips and tip usage in your lab by tenfold (10X).
On the plexWell Rapid Single Cell kit protocol for scRNA sequencing, simulation research was done at DISPENDIX. Based on the simulation data obtained, we were able to estimate the number of pipette tips required (Refer Table 1) to process one 96-well plate for plexWell Rapid Single Cell kit protocol by the conventional approach and with the I.DOT.
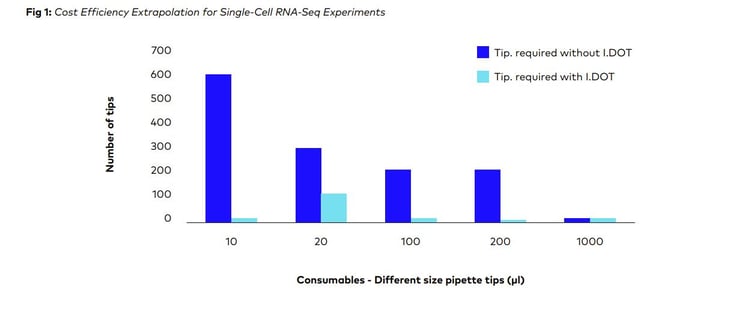
Additionally, it was also determined how much money and how many pipette tips could be saved by using this approach (Refer Fig: 2 & 3)
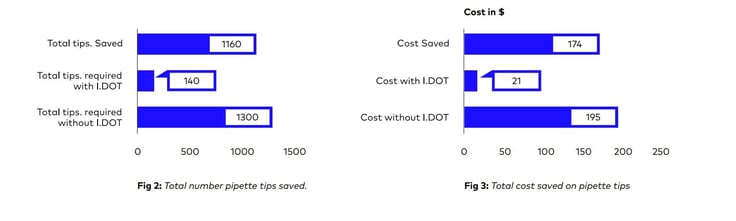
In conclusion, using I.DOT in scRNA seq. protocols minimizes consumable costs, allow you to miniaturize and automate the protocol, enable you to process multiple plates in a notably short period of time, eliminate cross-contamination issues, and improve the accuracy of your library preparation. Furthermore, reducing the use of plastic consumables in library preparation protocols, it contributes significantly to the
promotion of sustainable sequencing.
Check out the following articles for I.DOT application in NGS library preparation:
Optimizing NGS Library Prep
High Quality NGS Library Prep: Enhancing Understanding of Intra-Tissue Heterogeneity.
Book a demo today to learn more.