Next-generation sequencing (NGS), synthetic biology (SynBio), and innovative drug discovery techniques are essential for scientific research and development. These advancements have not only expanded our understanding of critical scientific processes but also facilitated groundbreaking developments in medicine, biotechnology, and environmental science. However, the complexity of assay development for these fields poses unique challenges, from streamlining NGS assays to ensuring accuracy in drug discovery and harnessing the potential of SynBio for practical applications.
Here, we delve into the intricacies of developing and validating assays across these fields, highlighting the pivotal role of emerging technologies and strategic methodologies in streamlining the process and enhancing the reliability and effectiveness of research outcomes.
Streamlining NGS Assay Development
NGS represents a pivotal advancement in DNA and RNA sequencing technologies, offering rapid, high-throughput, and cost-effective sequencing capabilities1. This technology enables the sequencing of millions of fragments simultaneously, providing insights into genetic variations and patterns critical for scientific discovery, diagnostics, and personalized medicine2–4. However, researchers face significant challenges in the development of robust NGS assays, including inefficient library preparation, low signal-to-noise ratios, and difficulties in achieving consistent results5–7.
Strategies to overcome these challenges include optimizing experimental designs, implementing rigorous quality control measures, and employing liquid handling automation7,8. Automated liquid handling, such as DISPENDIX's I.DOT Non-Contact Dispenser, streamlines NGS workflows by minimizing manual errors and variability in pipetting tasks, thus enhancing assay reliability and reproducibility (Fig. 1)9,10.
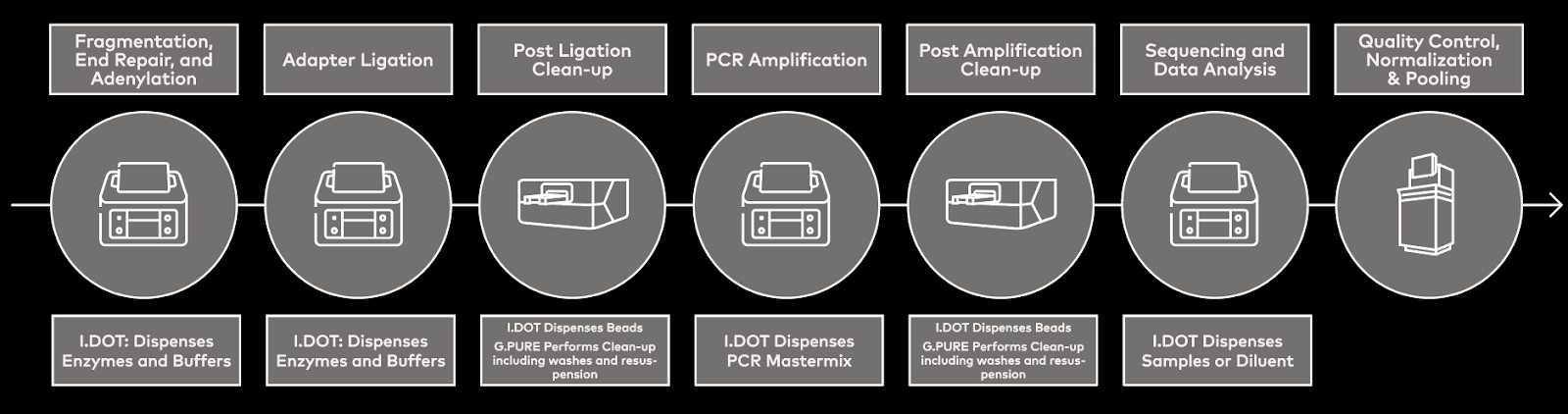
Figure 1. Workflow schematic showing where the I.DOT Liquid Handler and G.PURE Clean-up Device fit into the NGS library preparation workflow.
To learn more about techniques for streamlining NGS assay development, check out From Bench to Breakthrough: Streamlining NGS Assay Development Challenges for Success.

Assay Development and Validation for Drug Discovery
Developing new drugs is a complex and resource-intensive process11,12, and robust assay development and validation are critical for supporting the identification of promising therapeutic candidates. Assays, such as enzyme-linked immunosorbent assay (ELISA), enzyme activity assays, and cell viability assays, are utilized at various stages of drug discovery to evaluate compound libraries, characterize candidates, and optimize compounds based on their effects on cell health13–15.
Challenges in assay development include managing false positives/negatives, assay variability, and non-specific interactions, which can be mitigated through strategic design and validation practices16–18.
Emerging technologies, including microfluidic devices, biosensors, artificial intelligence, and automated liquid handling, play a significant role in enhancing assay development and validation19. These technologies, including the I.DOT Non-Contact Dispenser, improve assay throughput, precision, and reproducibility while reducing manual errors and sample volumes, thus streamlining drug discovery assays for better efficiency and accuracy.
To explore more top tips for assay development and validation for drug discovery, read Boosting Accuracy and Efficiency: Assay Development and Validation Process for Drug Discovery.
The Synthetic Biology Assay Development Pipeline
Synthetic biology is a broad field that applies engineering principles to biological system development20. The design-build-test-learn (DBTL) development pipeline demands a multifaceted technological scope, including DNA synthesis and sequencing, computational design, and omics technologies, to craft robust SynBio assays21. Key considerations for successful SynBio assay development include selecting the right biological components for desired functionality, adhering to modular design principles for system flexibility, and ensuring high-throughput screening compatibility for efficient testing22.
Advanced technologies play crucial roles in enhancing assay development: AI streamlines design, microfluidic devices allow precise control over experimental conditions, facilitating miniaturization and throughput, and automated liquid handling tools like the I.DOT Liquid Handler increases productivity, precision, and consistency (Fig. 2)23.
.png?width=600&height=338&name=alpha_I.DOT_.0AT%205%20(1).png)
Figure 2. The I.DOT Non-Contact Dispenser enables SynBio researchers to streamline their assay development processes for rapid, reproducible, and accurate workflows.
To delve deeper into SynBio assay development strategies, head to Streamline Your Research: Tips for Synthetic Biology Assay Development Technology.
Conclusion
In conclusion, assay development for NGS, drug discovery, and synthetic biology requires a delicate balance of innovation, precision, and strategic planning to overcome inherent challenges and achieve meaningful results.
The integration of advanced technologies such as automated liquid handling systems, AI, and microfluidics has been instrumental in addressing these challenges, offering new pathways to efficiency and accuracy. The continuous evolution of these tools and methodologies promises to further refine assay development processes, enabling scientists to push the boundaries of what is possible in genetic research, pharmaceuticals, and synthetic biology.
To learn more about how DISPENDIX’s I.DOT can accelerate your assay development pipelines for NGS, drug discovery, or SynBio, schedule a demo with one of our expert team members today.
References
- Milestones in Genomic Sequencing. Accessed March 13, 2024. https://www.nature.com/articles/d42859-020-00099-0
- Vandeputte M. The journey from next-generation sequencing to personalized medicine? The Biochemist. 2021;43(6):4-8. doi:10.1042/bio_2021_192
- Buermans HPJ, Den Dunnen JT. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta BBA - Mol Basis Dis. 2014;1842(10):1932-1941. doi:10.1016/j.bbadis.2014.06.015
- Fernandez-Marmiesse A, Gouveia S, Couce ML. NGS Technologies as a Turning Point in Rare Disease Research , Diagnosis and Treatment. Curr Med Chem. 2018;25(3):404-432. doi:10.2174/0929867324666170718101946
- Head SR, Komori HK, LaMere SA, et al. Library construction for next-generation sequencing: Overviews and challenges. BioTechniques. 2014;56(2):61-77. doi:10.2144/000114133
- Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19(5):269-285. doi:10.1038/nrg.2017.117
- Cheng C, Fei Z, Xiao P. Methods to improve the accuracy of next-generation sequencing. Front Bioeng Biotechnol. 2023;11:982111. doi:10.3389/fbioe.2023.982111
- Syed F, Grunenwald H, Caruccio N. Optimized library preparation method for next-generation sequencing. Nat Methods. 2009;6(10):i-ii. doi:10.1038/nmeth.f.269
- Socea JN, Stone VN, Qian X, Gibbs PL, Levinson KJ. Implementing laboratory automation for next-generation sequencing: benefits and challenges for library preparation. Front Public Health. 2023;11:1195581. doi:10.3389/fpubh.2023.1195581
- Hess JF, Kohl TA, Kotrová M, et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol Adv. 2020;41:107537. doi:10.1016/j.biotechadv.2020.107537
- Wouters OJ, McKee M, Luyten J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018. JAMA. 2020;323(9):844. doi:10.1001/jama.2020.1166
- Brown DG, Wobst HJ, Kapoor A, Kenna LA, Southall N. Clinical development times for innovative drugs. Nat Rev Drug Discov. 2022;21(11):793-794. doi:10.1038/d41573-021-00190-9
- Fu W, Chen Y, Wang K, et al. Repurposing FDA-approved drugs for SARS-CoV-2 through an ELISA-based screening for the inhibition of RBD/ACE2 interaction. Protein Cell. 2021;12(7):586-591. doi:10.1007/s13238-020-00803-w
- A. Haubrich B, C. Swinney D. Enzyme Activity Assays for Protein Kinases: Strategies to Identify Active Substrates. Curr Drug Discov Technol. 2016;13(1):2-15. doi:10.2174/1570163813666160115125930
- Larsson P, Engqvist H, Biermann J, et al. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci Rep. 2020;10(1):5798. doi:10.1038/s41598-020-62848-5
- Burt T, Button K, Thom H, Noveck R, Munafò M. The Burden of the “False‐Negatives” in Clinical Development: Analyses of Current and Alternative Scenarios and Corrective Measures. Clin Transl Sci. 2017;10(6):470-479. doi:10.1111/cts.12478
- Bajorath J. Evolution of assay interference concepts in drug discovery. Expert Opin Drug Discov. 2021;16(7):719-721. doi:10.1080/17460441.2021.1902983
- Gupta A, Gautam P, Wennerberg K, Aittokallio T. A normalized drug response metric improves accuracy and consistency of anticancer drug sensitivity quantification in cell-based screening. Commun Biol. 2020;3(1):42. doi:10.1038/s42003-020-0765-z
- Ramadan Q, Perinelli DR, Orian L, Baratchi S. Editorial: Innovative approaches in drug discovery and development. Front Med Technol. 2023;5:1206088. doi:10.3389/fmedt.2023.1206088
- Synthetic Biology. Accessed April 5, 2024. https://www.genome.gov/about-genomics/policy-issues/Synthetic-Biology
- Kitano S, Lin C, Foo JL, Chang MW. Synthetic biology: Learning the way toward high-precision biological design. PLOS Biol. 2023;21(4):e3002116. doi:10.1371/journal.pbio.3002116
- Garner KL. Principles of synthetic biology. Essays Biochem. 2021;65(5):791-811. doi:10.1042/EBC20200059
- Ortiz L, Pavan M, McCarthy L, Timmons J, Densmore DM. Automated Robotic Liquid Handling Assembly of Modular DNA Devices. J Vis Exp. 2017;(130):54703. doi:10.3791/54703