FLASH-seq (FS) is a new single-cell RNA sequencing method presented in the paper Fast and highly sensitive full-length single-cell RNA sequencing using FLASH-seq by researchers from the Institute of Molecular and Clinical Ophthalmology Basel and the Chemical Biology and Therapeutics, Novartis Institutes for Biomedical Research (Fig. 1). that offers significant improvements over existing techniques. FS boasts higher sensitivity (meaning it catches a wider range of genes) and requires less hands-on time (roughly 4.5 hours) compared to its popular predecessor, Smart-seq3. This faster protocol is also easily automated and miniaturized, reducing resource demands.

Figure 1. Screenshot of the paper Fast and highly sensitive full-length single-cell RNA sequencing using FLASH-seq in the publication Nature Biotechnology.
FS incorporates unique molecular identifiers (UMIs) for accurate gene expression quantification while minimizing artifacts.
The I.DOT Liquid Handler was used in lysis buffer preparation and RT-PCR [reverse transcription (RT) and cDNA preamplification (polymerase chain reaction)] reactions for FS and FS-LA (FLASH-seq low amplification) (Fig. 2).
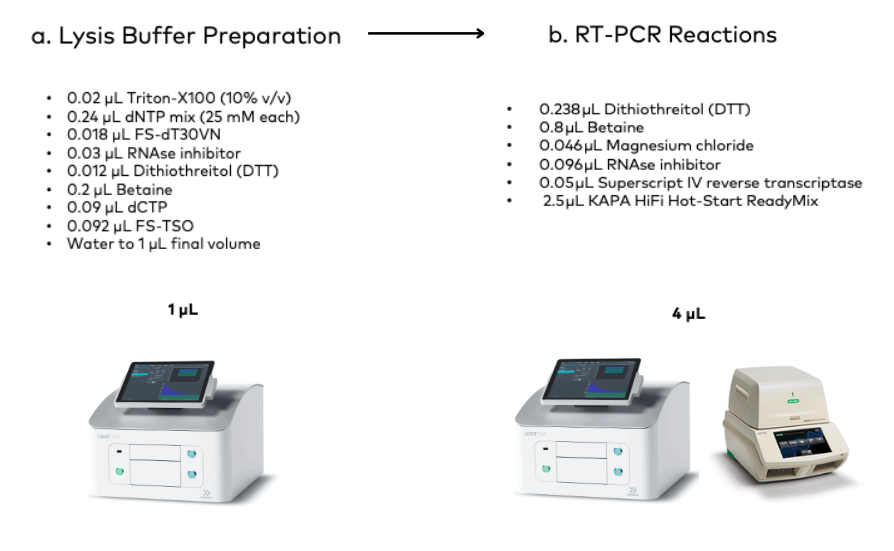
Figure 2. Breakdown of lysis buffer preparation and RT-PCR reactions using the I.DOT Liquid Handler.
Lysis Buffer Preparation
The FLASH-seq and FLASH-seq low amplification protocols involve preparing a buffer that lyses the cells and preserves their RNA for sequencing (Fig. 2a). This buffer is then dispensed into individual wells of a 384-well plate, where each well holds a single cell.
Preparation and Storage
- The lysis buffer components are mixed together.
- The buffer is dispensed into individual wells of a 384-well plate using the I.DOT Liquid Handler.
- The plate is sealed and stored at -20°C until needed.
RT-PCR Reactions
1. Plates containing lysed cell samples were thawed from -80°C and warmed up at 72°C for 3 minutes.
2. Using the I.DOT Non-Contact Dispenser, 4 µL of RT-PCR mix were added to each well (Fig. 2b). This mix contains key components for reverse transcription (Fig. 2b)
3. The plate was then incubated at 50°C for 1 hour.
4. Following reverse transcription, the reaction undergoes an initial denaturation step at 98°C for 3 minutes. This ensures all cDNA molecules are single-stranded for efficient amplification.
5.PCR amplification then occurs through repeated cycles of denaturation, annealing, and extension.
FS promises to be a powerful tool for studying gene expression across numerous samples with exquisite detail, making it a valuable asset for diverse research applications.

Accelerate your scRNA-seq Research
Harness the power of the I.DOT Liquid Handler to:
-
Effortlessly dispense nanoliter volumes: Ensure precise reagent delivery with exceptional accuracy and reproducibility.
-
Streamline multi-step workflows: Automate complex processes for lysis buffer and RT-PCR mix additions, saving you valuable time and effort.
-
Minimize sample loss and contamination: Protect precious samples with the I.DOT's exceptional contact-free dispensing technology.
-
Enhance experiment reproducibility: Achieve consistent results across your experiments with the I.DOT's unparalleled precision.
Ready to experience the I.DOT advantage? Download the I.DOT brochure and learn how the I.DOT Non-Contact Dispenser can revolutionize your single-cell research.
Don't let manual pipetting hold you back. Embrace automation and unlock the full potential of your scRNA-seq experiments with the I.DOT Non-Contact Dispenser.
References
Hahaut, V., Pavlinic, D., Carbone, W. et al. Fast and highly sensitive full-length single-cell RNA sequencing using FLASH-seq. Nat Biotechnol 40, 1447–1451 (2022). https://doi.org/10.1038/s41587-022-01312-3