Cell-free protein synthesis (CFPS) is a versatile technology for synthetic biology and bioengineering that allows the production of proteins outside of living cells. Cell extracts containing the essential cellular components for protein synthesis, such as ribosomes, tRNAs, cofactors, and enzymes, are used in CFPS, in contrast to conventional techniques that rely on cellular systems. Since these components are supplied as cell lysates, it is difficult to regulate their concentrations, which leads to an inefficient cell-free system.
By applying artificial intelligence to vary one compound concentration at a time in their investigation of cell-free buffer compositions, scientists were able to achieve a 34-fold increase in protein production for in vitro ribosome assemblies. However, developing a lysate-specific optimization strategy for the composition of the cell-free buffer to maximize protein production, is a significant challenge as it depends on the optimization of reaction conditions and, consequently, on experiment design flexibility. The researchers also used an acoustic liquid handler that is slower, less accurate, with lower resolution, and more costly compared to the liquid handling solution that we present below.
Breaking CFPS Barriers with I.DOT Technology
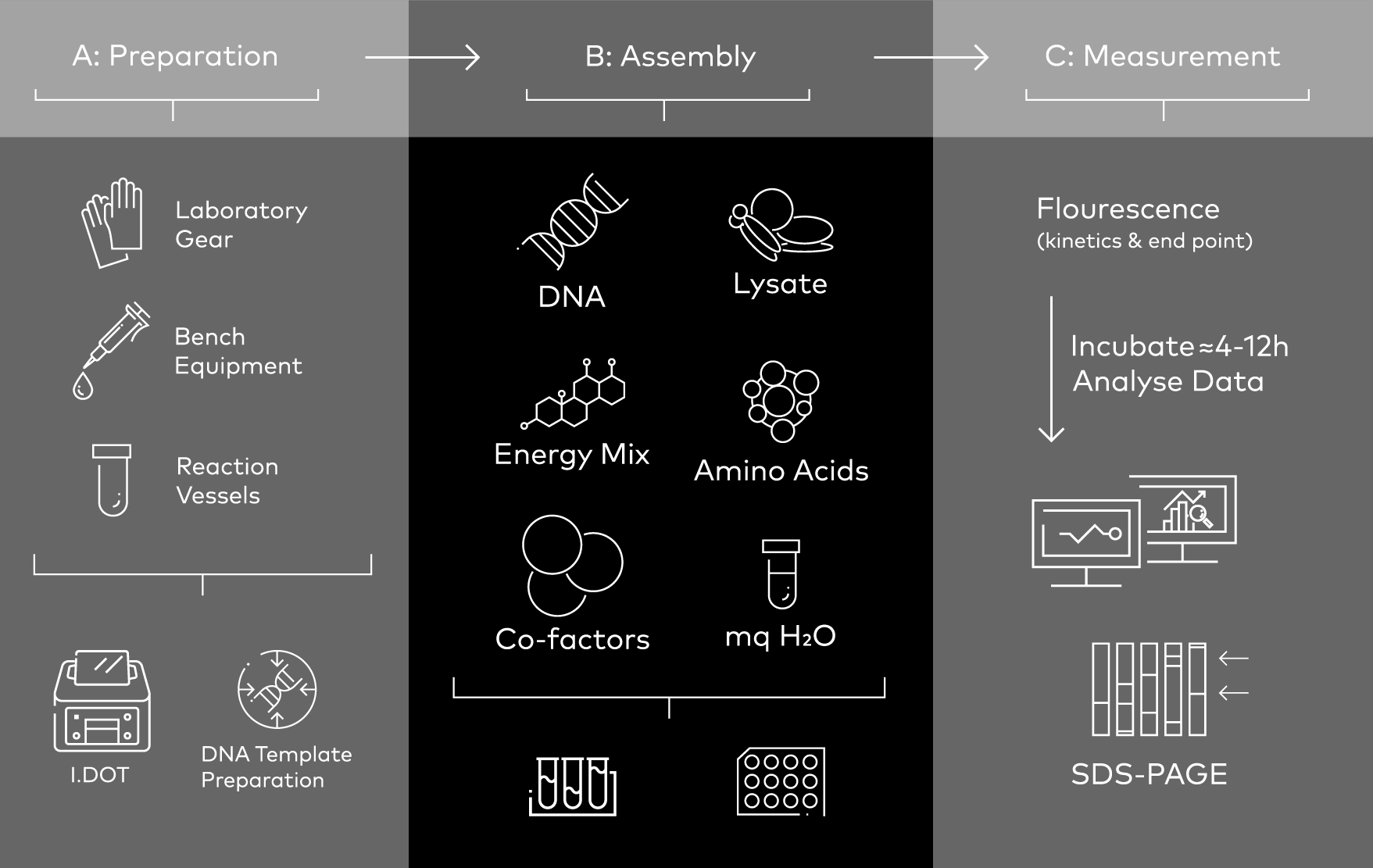
Figure 1. Cell-free protein synthesis (CFPS) workflow: the I.DOT Liquid Handler could be an essential part of the template preparation and reagent assembly steps of CFPS assays. Image modified from Garenne et al. (2021).

The integration of the I.DOT Liquid Handler with cell-free protein synthesis (CFPS) workflows would enhance the scalability, precision, and automation of the protein synthesis processes (Fig. 1). It not only makes the experimental workflow more efficient by lowering both the dispense time and overall reaction cost, but it also would make it possible to optimize conditions with an unprecedented level of accuracy and improving experiment reproducibility.
When it comes to applications like protein synthesis for research, biotechnology, or drug discovery—where high-throughput screening (HTS), automation, and optimization are essential—this integration is especially valuable. Other benefits of using the I.DOT Non-Contact Dispenser in the assays for CFPS workflows include the following:
Flexibility
The I.DOT Liquid Handler is programmed to dispense specific reagents, such as cell extract, DNA/RNA templates, amino acids, energy sources, and cofactors, into the reaction mixture. Because they allow for the simultaneous handling of numerous reagents and samples, liquid handlers provide flexibility in the design of intricate experiments. When working with a variety of experimental setups and variations, this flexibility is essential.
Optimization of Reaction Conditions
Cell-free gene expression reactions often require optimization of conditions such as temperature, buffer composition, and reagent concentrations. The I.DOT Non-Contact Dispenser allows for systematic and precise exploration of these parameters, facilitating the identification of optimal conditions for protein expression.
Miniaturization and Resource Efficiency
Miniaturizing reactions with the I.DOT Non-Contact Dispenser reduces the amounts of reagents required, making experiments more resource-efficient.
Integration with Analytical Techniques
To monitor protein synthesis in real time or assess the quality of synthesized proteins, the I.DOT Non-Contact Dispenser can be integrated with a variety of other analytical tools, like mass spectrometers or fluorometers.
Transform your Protein Synthesis Systems
In the pursuit of excellence, time and precision are paramount. Our team and our customers have embraced Immediate Drop-On-Demand Technology (I.DOT), propelling researchers into a new era of control and accuracy in dispensing reaction components. This isn't just about assays; it's about precision engineering for results you can trust.
Curious about how the I.DOT Liquid Handler can elevate your protein synthesis assays? Download the I.DOT brochure to see firsthand how precision and speed can redefine your laboratory workflows.
References
Borkowski, O., Koch, M., Zettor, A., Pandi, A., Batista, A.C., Soudier, P. and Faulon, J.L., 2020. Large scale active-learning-guided exploration for in vitro protein production optimization. Nature communications, 11(1), p.1872.
Carlson, E.D., Gan, R., Hodgman, C.E. and Jewett, M.C., 2012. Cell-free protein synthesis: applications come of age. Biotechnology advances, 30(5), pp.1185-1194.
Caschera, F., Karim, A.S., Gazzola, G., d’Aquino, A.E., Packard, N.H. and Jewett, M.C., 2018. High-throughput optimization cycle of a cell-free ribosome assembly and protein synthesis system. ACS synthetic biology, 7(12), pp.2841-2853.
Garenne, D., Haines, M.C., Romantseva, E.F., Freemont, P., Strychalski, E.A. and Noireaux, V., 2021. Cell-free gene expression. Nature Reviews Methods Primers, 1(1), p.49.