Next-generation sequencing (NGS) is an advanced, high-throughput DNA and RNA sequencing technique. Its development represented a significant advancement in sequencing technologies, making it possible for researchers to sequence DNA and RNA far more rapidly and cost-effectively than the traditional Sanger sequencing method1. In addition, NGS supports the parallel sequencing of millions of fragments, providing a vast depth of data that can uncover genetic variations, mutations, and patterns within a single assay2.
NGS has a broad range of applications, including identifying genetic mutations associated with diseases, mapping genetic populations, and tracing the evolution of species. Its ability to provide rapid, accurate, and detailed genomic information has driven scientific discovery, rare disease diagnostics, and personalized medicine3–5.
Despite its potential, developing robust NGS assays poses significant challenges, including issues related to library preparation efficiency, signal-to-noise ratio, and achieving consistent and reproducible results. We explore these assay development challenges and outline some strategies for success.
Understanding NGS Assay Development Challenges
The high sensitivity of NGS assays means that it is essential that workflows are designed with accuracy and precision in mind. Even the slightest fluctuation in assay setup can lead to massive variability in the results, making it challenging to differentiate genuine biological variation from procedural artifacts7. Some common assay development challenges commonly faced by researchers include:
1. Library Preparation Inefficiency
NGS library prep is notoriously tedious, time-consuming, and prone to error. Thus, there is no wonder it is often deemed the bottleneck of NGS. Ideally, library prep should result in a library that accurately reflects the input DNA. Inefficient library preparation poses the risk of over- or under-representing certain DNA regions, which can lead to inaccurate sequencing results8.
2. Low Signal-to-Noise Ratio
In NGS, a high signal-to-noise ratio is essential for distinguishing true genetic variants from sequencing errors and, consequently, for facilitating accurate interpretation9. Errors or inefficiencies in library prep, sequencing errors, and low-quality input material can cause low signal-to-noise ratios6.
3. Difficulty in Achieving Consistent Results
Achieving consistent NGS results is pivotal for the reliability and reproducibility of the resulting research10. However, several factors inherent to the sequencing workflow, sample variability, and data analysis processes can challenge this consistency and lead to unreliable results. Inconsistencies can manifest as variations in sequence coverage, discrepancies in variant calling, or differences in quality across runs or samples.
Strategies for Overcoming NGS Assay Development Challenges
Overcoming NGS assay development challenges demands a comprehensive approach that encompasses all stages of the NGS workflow, from experimental design to data normalization and QC10.
First, the experimental design, including sample selection and handling processes, should be well thought out and optimized for success. This includes developing and adhering to standardized protocols for sample preparation, library construction, and sequencing11. The library preparation process should be carefully optimized, including reagent, input material, and tool selection, to reduce artifacts and maximize signal-to-noise ratios12.
Implementing rigorous quality control measures, including internal controls and reference materials, at each step of the NGS workflow, from sample collection and library preparation to sequencing and data analysis, helps identify and mitigate sources of variability11.
Finally, applying appropriate normalization techniques and adopting a standardized bioinformatics pipeline can help correct for any technical variability and improve the consistency of data interpretation across different datasets13.

The Role of Liquid Handling Automation in Overcoming NGS Assay Development Challenges
Many of the most commonly experienced NGS assay development challenges are borne out of variability in the assay setup, library prep, and clean-up stages rather than the sequencing itself. Considering this, automating these processes represents a promising strategy for overcoming NGS assay development challenges and obtaining more accurate, reliable, and reproducible results14.
Integrating automated liquid handlers into NGS workflows can allow users to automate the essential but repetitive and labor-intensive pipetting tasks integral to the NGS workflow, such as DNA/RNA extraction, library preparation, and sample normalization15. Not only does liquid handling automation streamline the workflow, but it can also minimize the risk of cross-contamination and reagent waste, leading to cost-effective, consistent, and high-quality NGS data.
DISPENDIX’s I.DOT Liquid Handler is a non-contact liquid handling tool that supports rapid, low-volume liquid handling for 96- and 384-well plates. Moreover, it can be integrated with a range of other tools, such as the G.PURE Clean-up Device, into the G.STATION NGS Workstation for complete automation of NGS library prep (Fig. 1).
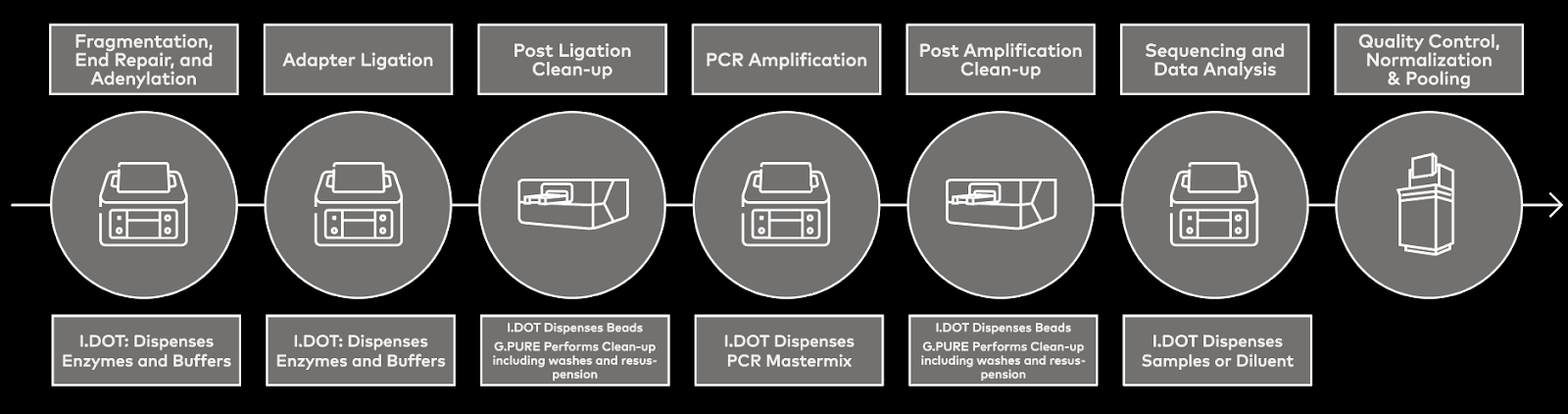
Figure 1. Workflow schematic showing where the I.DOT Liquid Handler and G.PURE Clean-up Device fit into the NGS library preparation workflow.
Conclusion
NGS is a revolutionary sequencing technology that enables rapid, high-throughput, cost-effective DNA and RNA sequencing. Despite its transformative potential, developing robust NGS assays presents significant challenges, including library preparation inefficiency, low signal-to-noise ratio, and difficulty achieving consistent results. Overcoming these assay development challenges through strategies such as rigorous quality control, optimized experimental design, and the adoption of liquid handling automation like DISPENDIX's I.DOT Non-Contact Dispenser is crucial for achieving consistent, reliable, and clinically meaningful NGS data.
The DISPENDIX team has worked through the miniaturization of even the most complex NGS workflows and can help you to scale volumes as low as 1/10th the manufacturers recommended SOP without compromising data quality, making DISPENDIX the perfect partner NGS assay development partner. Book your free demo today!
References
- Milestones in Genomic Sequencing. Accessed March 13, 2024. https://www.nature.com/articles/d42859-020-00099-0
- Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child - Educ Pract Ed. 2013;98(6):236-238. doi:10.1136/archdischild-2013-304340
- Vandeputte M. The journey from next-generation sequencing to personalized medicine? The Biochemist. 2021;43(6):4-8. doi:10.1042/bio_2021_192
- Fernandez-Marmiesse A, Gouveia S, Couce ML. NGS Technologies as a Turning Point in Rare Disease Research , Diagnosis and Treatment. Curr Med Chem. 2018;25(3):404-432. doi:10.2174/0929867324666170718101946
- Brittain HK, Scott R, Thomas E. The rise of the genome and personalised medicine. Clin Med Lond Engl. 2017;17(6):545-551. doi:10.7861/clinmedicine.17-6-545
- Buermans HPJ, Den Dunnen JT. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta BBA - Mol Basis Dis. 2014;1842(10):1932-1941. doi:10.1016/j.bbadis.2014.06.015
- Bacher U, Shumilov E, Flach J, et al. Challenges in the introduction of next-generation sequencing (NGS) for diagnostics of myeloid malignancies into clinical routine use. Blood Cancer J. 2018;8(11):113. doi:10.1038/s41408-018-0148-6
- Head SR, Komori HK, LaMere SA, et al. Library construction for next-generation sequencing: Overviews and challenges. BioTechniques. 2014;56(2):61-77. doi:10.2144/000114133
- Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19(5):269-285. doi:10.1038/nrg.2017.117
- Cheng C, Fei Z, Xiao P. Methods to improve the accuracy of next-generation sequencing. Front Bioeng Biotechnol. 2023;11:982111. doi:10.3389/fbioe.2023.982111
- Endrullat C, Glökler J, Franke P, Frohme M. Standardization and quality management in next-generation sequencing. Appl Transl Genomics. 2016;10:2-9. doi:10.1016/j.atg.2016.06.001
- Syed F, Grunenwald H, Caruccio N. Optimized library preparation method for next-generation sequencing. Nat Methods. 2009;6(10):i-ii. doi:10.1038/nmeth.f.269
- Roy S, Coldren C, Karunamurthy A, et al. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines. J Mol Diagn. 2018;20(1):4-27. doi:10.1016/j.jmoldx.2017.11.003
- Socea JN, Stone VN, Qian X, Gibbs PL, Levinson KJ. Implementing laboratory automation for next-generation sequencing: benefits and challenges for library preparation. Front Public Health. 2023;11:1195581. doi:10.3389/fpubh.2023.1195581
- Hess JF, Kohl TA, Kotrová M, et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol Adv. 2020;41:107537. doi:10.1016/j.biotechadv.2020.107537